FAFDrugs4




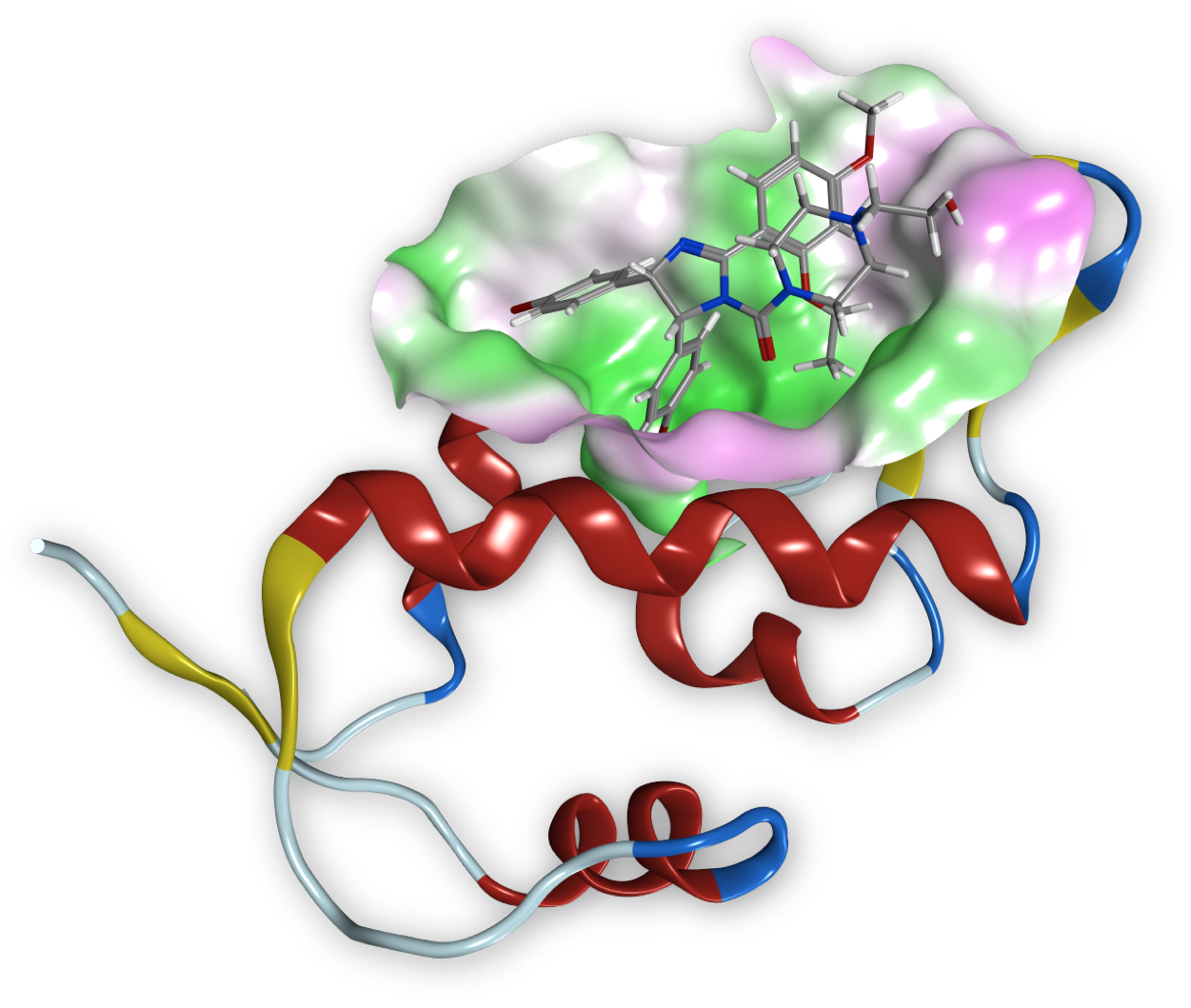
Protein-protein interactions (PPIs) are essential to life and various diseases states are associated with aberrant PPIs. Therefore significant efforts are dedicated to this new class of therapeutic targets, see beside, the structure of MDM2 co-crystallized with Nutlin-2 which inhibit MDM2-p53 binding. Even though it might not be possible to modulate the estimated 650,000 PPIs that regulate human life with drug-like compounds, a sizeable number of PPI should be druggable.
Aberrant PPIs contribute to most disease states and therefore represents a highly populated class of essentially untouched targets for drug discovery. We advocate that a possible solution to this conundrum is to mine relevant drug-like PPI inhibitors and define a dedicated profile through the use of appropriate chemoinformatics and machine learning tools.
To this end, we designed a decision tree [50],[51] constructed with the two Dragon descriptors Ui and RDF070m [52], trained on a data set composed of true PPI inhibitors and non-PPI inhibitors and validated on the experimental screening results of 11 representative protein-protein interactions. RDF070m is a Radial Distribution Function (RDF(r)) descriptor weighted by the atomic masses using a sphere radius r of 7Å as the associated probability distribution function and known as a shape descriptor. Ui is the unsaturation index, directly correlated to the number of multiple bonds: double, triple and most predominantly aromatic bonds.
The decision tree was transposed into a computer program named PPI-HitProfiler [51], that can be used as a filter to enrich chemical libraries in putative iPPI.