FAFDrugs4




According to Baell et al. [23] , PAINS moieties, standing for Pan Assay Interference Compounds, are compounds that appear as frequent hitters (promiscuous compounds) in many biochemical high throughput screens.
Whilst 90% of the compounds were either inactive or were only a hit in a single assay, a significant number of the compounds (0.39%) were active in all 6 assays tested. Exploring the structures that appeared promiscuous they were able to identify certain structural types a selection. Among others, Quinones-like (left below) or Rhodanines-like (right below) compromise the structures that are widely reported to be false positives.
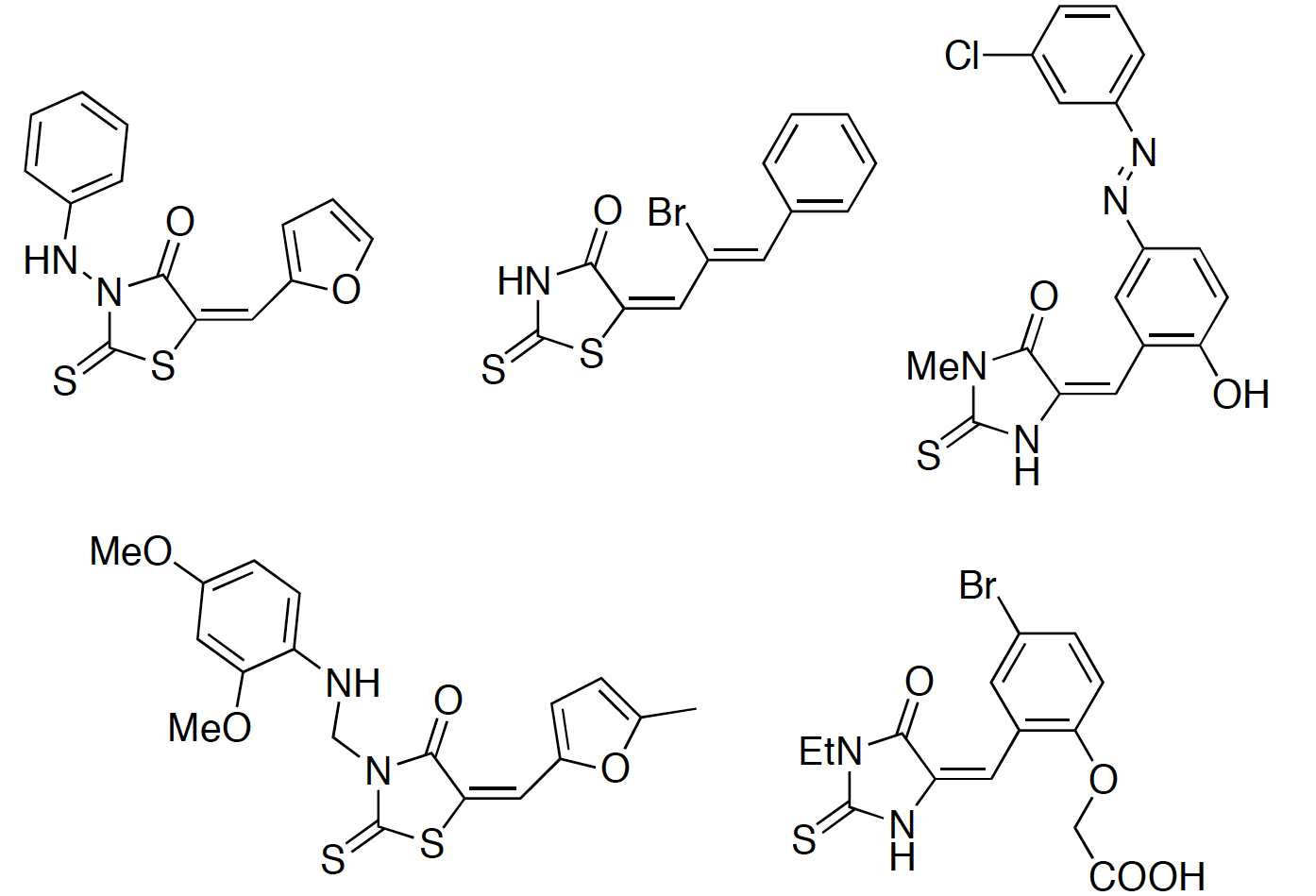
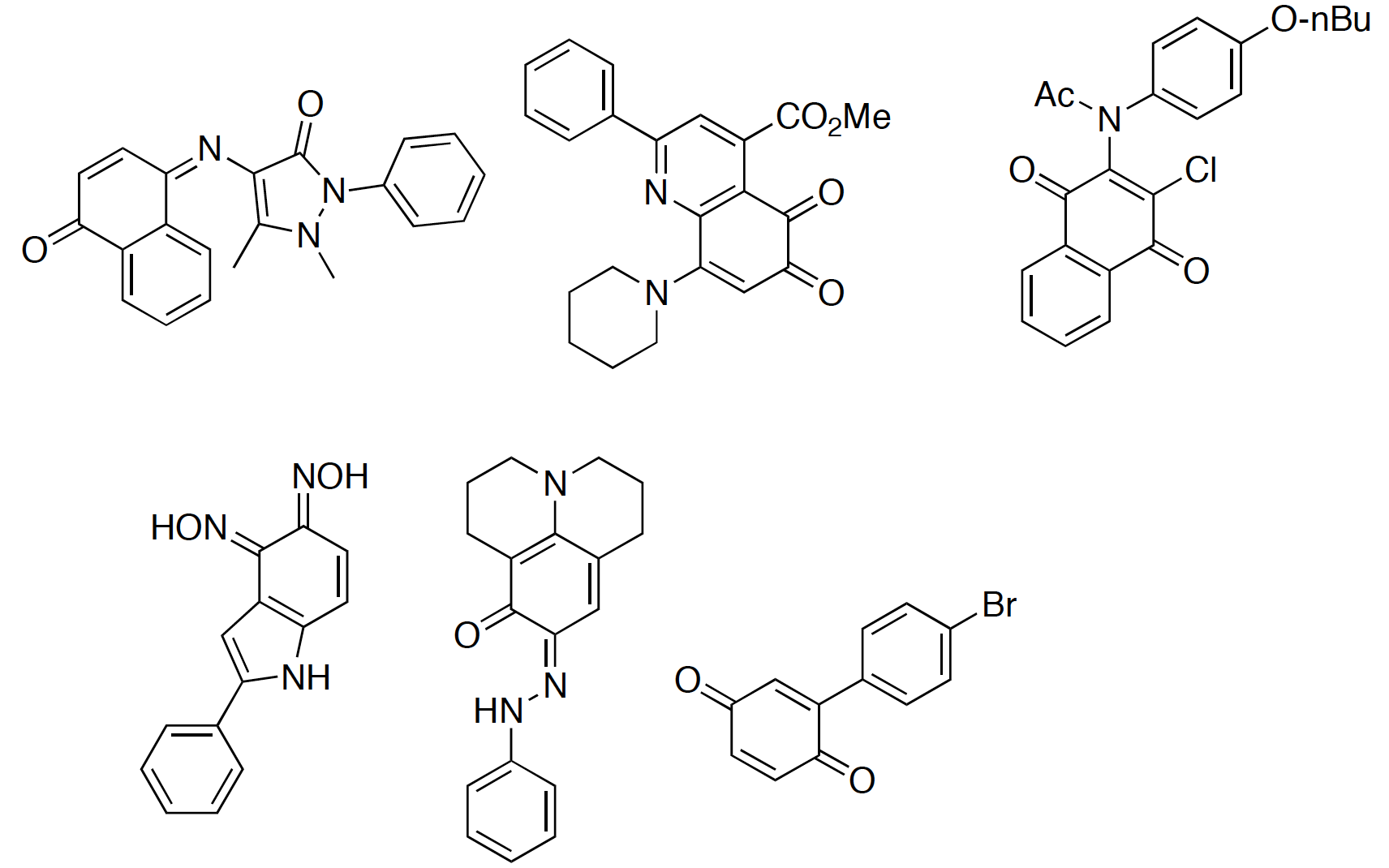
The compounds identified by such substructural features are not recognized by filters commonly used to identify reactive compounds. Even though these substructural features were identified using only one assay detection technology, such compounds have been reported to be active from many different assays. In fact, these compounds are increasingly prevalent in the literature as potential starting points for further exploration, whereas they may not be.
Published filters A, B, and C are applied in FAF-Drugs4, and are inspired from the SMARTS that Dr. Guha derived [42] from the original sln provided in reference [23]. Those had to be modified so as to reproduce results of a control set (see Supplementary Data Table 1-2).
Recent discussions about PAINS can be found in "Locating Sweet Spots for Screening Hits and Evaluating Pan-Assay Interference Filters from the Performance Analysis of Two Lead-like Libraries. Mok NY et al. J Chem Inf Model. 2013
We would like to thank C. GAGEAT who contributed to their optimization.